New Protein Language Model by Former Meta Scientists - Aiowio
A team of former Meta scientists has unveiled a groundbreaking model capable of generating proteins in a manner similar to how chatbots produce text. This revolutionary model, named ESM3, was introduced by EvolutionaryScale in June. Founded by ex-Meta researchers with a specialization in AI models for biology, EvolutionaryScale described ESM3 as “the first generative model for biology.”
Model Training and Functionality
The ESM3 model has been trained on the sequence, structure, and function of over 2.7 billion proteins, which allows it to generate novel proteins based on specific prompts. This advancement marks a significant step towards creating programmable tools for the field of biology.
In an interview with Nature, Alexander Rives, the chief scientist at EvolutionaryScale and former lead of Meta’s “AI protein team,” emphasized the aim to make biology programmable through the development of such tools.
Investment and Recognition
In a notable development, EvolutionaryScale secured $142 million in a seed round in June, with investments coming from Lux Capital, Amazon Web Services, Nat Friedman, Daniel Gross, and NVentures, the venture capital arm of Nvidia. Lux Capital’s Josh Wolfe likened EvolutionaryScale’s achievement to a pivotal moment akin to the ChatGPT breakthrough in the realm of biology.
During their tenure at Meta, Rives and his team compiled a database of over 600 million protein structures primarily focused on drug development. Previous iterations of the ESM model, such as ESMFold, leveraged a large language model trained on biological data to forecast protein structures. However, the team was disbanded in 2023 as part of Meta’s restructuring efforts to prioritize commercial AI products.
Recent Developments and Impact
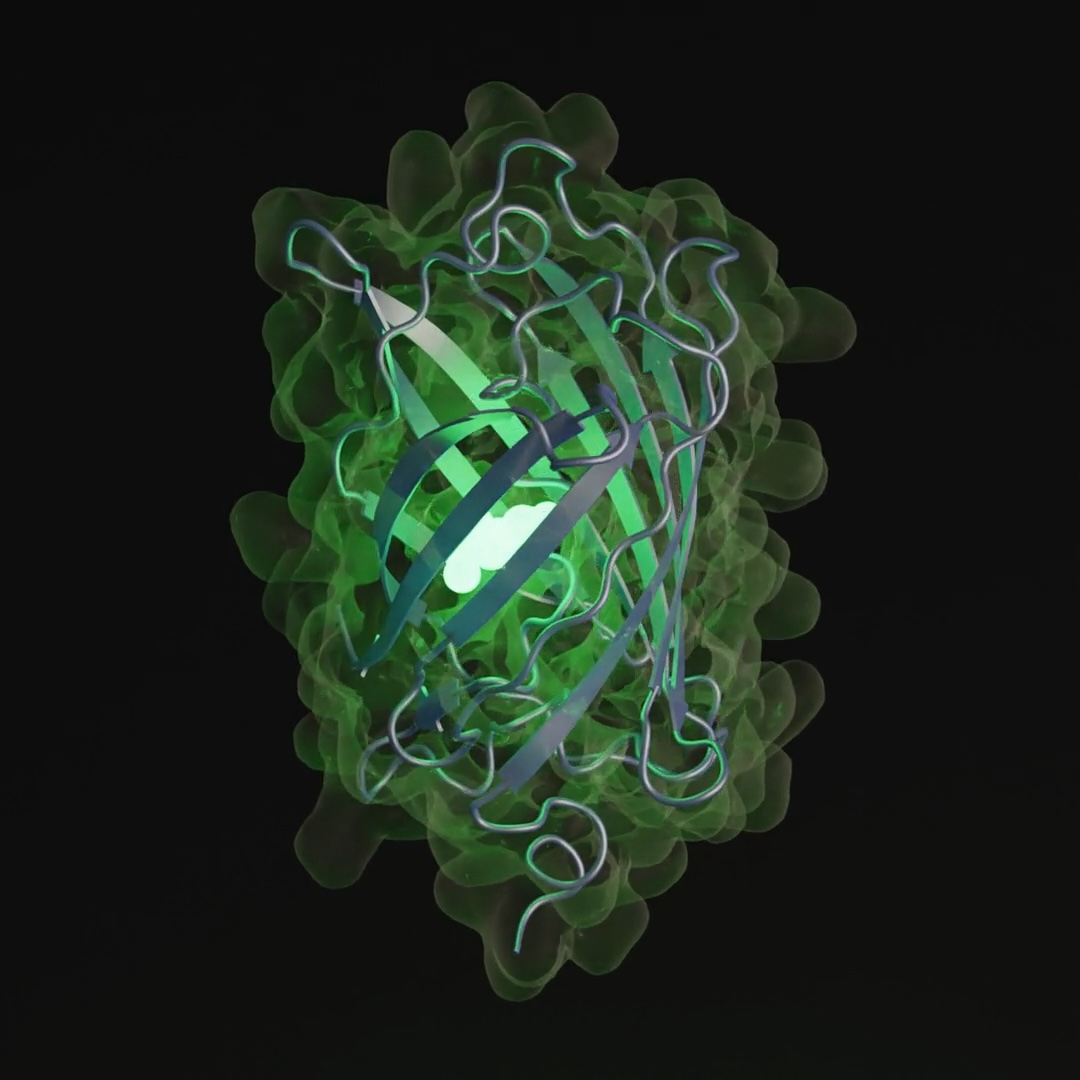
EvolutionaryScale recently teased a forthcoming paper showcasing ESM3’s ability to generate new green fluorescent proteins (GFPs). The generated sequence exhibits a 58% dissimilarity to the nearest known fluorescent protein, replicating the characteristics of proteins responsible for the bioluminescence observed in jellyfish and coral. EvolutionaryScale hailed this feat as a simulation of over 500 million years of evolutionary progress, reflecting the natural rate of GFP diversification.
.png)










